Section outline
-
Artemis Rumbou, Susanne von Bargen, Kira Köpke, Carmen Büttner
Humboldt-Universität zu Berlin, Albrecht Daniel Thaer-Institute, Division Phytomedicine, Berlin, Germany


-
-
Urban pest & disease
Mosaic disease of maple
Pathogen(s)
maple mottle-associated virus (Emaravirus aceris), maple emaravirus 2 (MEV-2), maple emaravirus 3 (MEV-3)
-
-
-
Mosaic, mottle, leaf deformation. We assume that the mottle symptoms exhibited in infected leaves may lead to reduced photosynthetic capacity and, consequently, to deterioration in the health of the trees (Fig. 1).
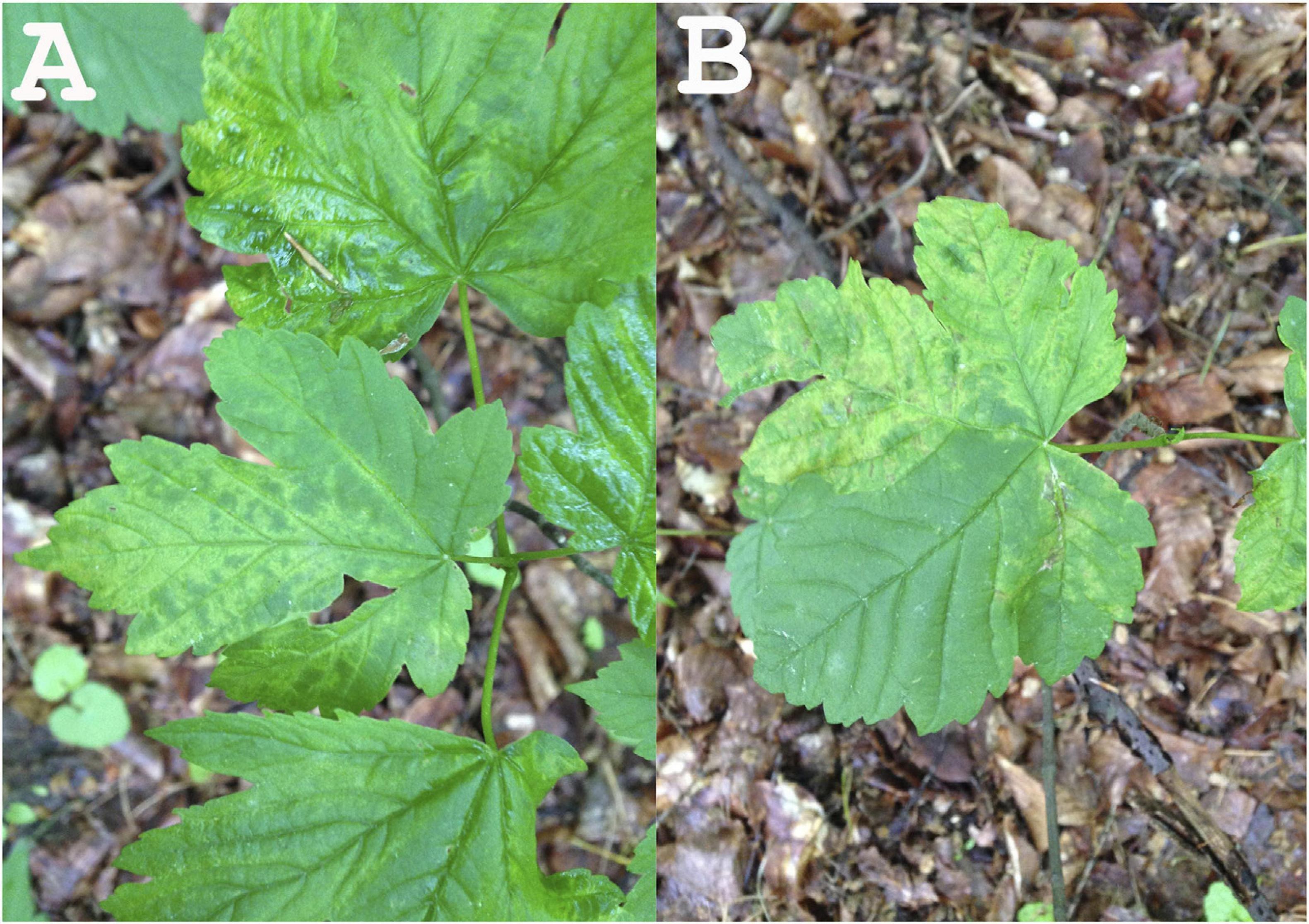
Figure 1. Leaf symptoms of the sycamore tree infected with emaravirus aceris (A: leaf mottle; B: leaf mottle and deformation).
-
-
-
Acer pseudoplatanus
-
-
-
The use of tested virus-free plant material is highly recommended when following the demand of planting healthy trees. This is even more important in urban ecosystems, where trees grow under adverse environmental conditions that can significantly impact their health (Bandte et al., 2022). Other viruses have also been reported to infect Acer spp. (Büttner et al. 2023) and induce virus-like symptoms on leaves, reliable diagnosis of MaMaV should be done by molecular means. Especially in the case of the partial genomes of the recently identified novel emaraviruses (MEV-2, MEV-3) related to MaMaV in mosaic-diseased maple trees, virus-specific RT-PCR from symptomatic leaf material should be applied in combination with sequencing, to discriminate between different species or strains by these emaraviruses.
-
-
-
Not regulated within the EU
-
-
-
Germany (Berlin, Brandenburg, Bavaria), Italy (Bozen)
The disease was first confirmed in Germany to be associated with a novel emaravirus maple mottle-associated virus (MaMaV), but virus-like symptoms have been observed and reported earlier in maples not only in Germany (Bandte et al., 2008; Büttner et al., 2013), but also in Romania (Ploiaie and Macovei, 1968), Hungary (Szirmai, 1972), Turkey (Erdiller, 1986), North America (Brierley, 1944), United Kingdom (Cooper, 1979). Symptom descriptions such as “mosaic,” “chlorotic mottle,” “yellow-mottle,” “ring-mottle,” and “mosaic mottling with chlorotic spots” were used by earlier researchers. Whether the newly identified viruses might be associated with the symptoms described in these earlier studies remains to be determined, given in particular that electron-dense structures known as double-membrane-bound bodies with diameters differing within the range of 80–200 nm and typical of emaraviruses (Mielke-Ehret and Mühlbach, 2012) have never been reported in earlier electron microscopy samples from symptomatic maples, while more typical plant virus particles sometimes have.
-
-
-
The genome of MaMaV is composed of six negative single-stranded RNA molecules. complete sequence of six genomic RNA segments were obtained (RNA1: 7,074 nt; RNA2: 2,289 nt; RNA3: 1,525 nt; RNA4: 1,533 nt, RNA5: 1,825 nt; RNA6: 1,179 nt) (Fig. 2). The full-length genomic sequences of the RNA segments are deposited in GenBank under accession numbers MT879190–MT879195. All six RNAs share a fully conserved stretch of 13 nt at their 5′ and 3′ termini.
ORF1 predicted on RNA1 is 2,305 aa-long and the encoded protein shows significant aa identity with the viral replicase of 21 known emaraviruses (ORF1: nt positions 6,966 – 49). The putative RNA polymerase exhibits highest aa identity with that of rose rosette virus (Accession number: QHZ99251.1; 74.4% aa identity).
ORF2 encoded on RNA2 is predicted to encode a protein of 646 aa (nt positions 1,996 – 56). The encoded protein shows aa identity with 21 other emaraviruses. The highest aa identity is with the glycoprotein precursor (GPP) of rose rosette virus (Accession number: QID76023.1; aa identity: 56.7%).
ORF3 encoded on RNA3 is predicted to encode a protein of 300 aa (nt positions 104 – 1,006) with highest BLASTp aa identity with the nucleocapsid proteins (NP) of the 21 known emaraviruses. The ORF3-encoded protein shows highest aa identity with the NP protein of Actinidia emaravirus 2 (Accession number: QEE82888.1; aa identity: 55.6%).
ORF4 encoded on RNA4 is predicted to encode a protein of 363 aa (nt positions 1,179 – 88) which shows very variable BLASTp aa identity levels with the movement proteins identified from 20 emaraviruses. It exhibits highest aa sequence identity with the movement protein of fig mosaic virus (Accession number: BAM13817.1; aa identity: 65.2%).
The fifth predicted ORF is encoded on RNA5 and is predicted to encode a protein of 373 aa (nt positions 1,526 – 93). The hypothetical protein encoded by RNA5 shows sequence homology with the corresponding protein detected from eight emaraviruses. This putative protein shares highest identity with the P5 protein of rose rosette virus (Accession number: QIB98219. 1; aa identity: 44.8%), a protein of unknown function. The rest of the emaraviruses do not have a homologous protein or such has not been identified yet.
Finally, the sixth ORF, encoded on RNA6 (nt positions 766 – 71), produces a 231 aa-long protein with 35–45% aa identity with that of other emaraviruses. The highest aa identity is with the P6 protein from Actinidia emaravirus 2 (Accession number: QEE82891.1; aa identity: 44.2%). The novel virus is found to have a homologous protein to the one encoded by RNA6 segment of only eight from the already known emaraviruses.
To summarize, MaMaV is phylogenetically related to the emaraviruses currently represented in the GenBank belonging to the novel virus species Emaravirus aceris. It clustering with the largest clade, the “clade A” in the Emaravirus genus, which includes rose rosette virus (Emaravirus rosae), Actinidia emaravirus 2 (Emaravirus kiwii), and fig mosaic virus (Emaravirus fici) (Rehanek et al. 2022).
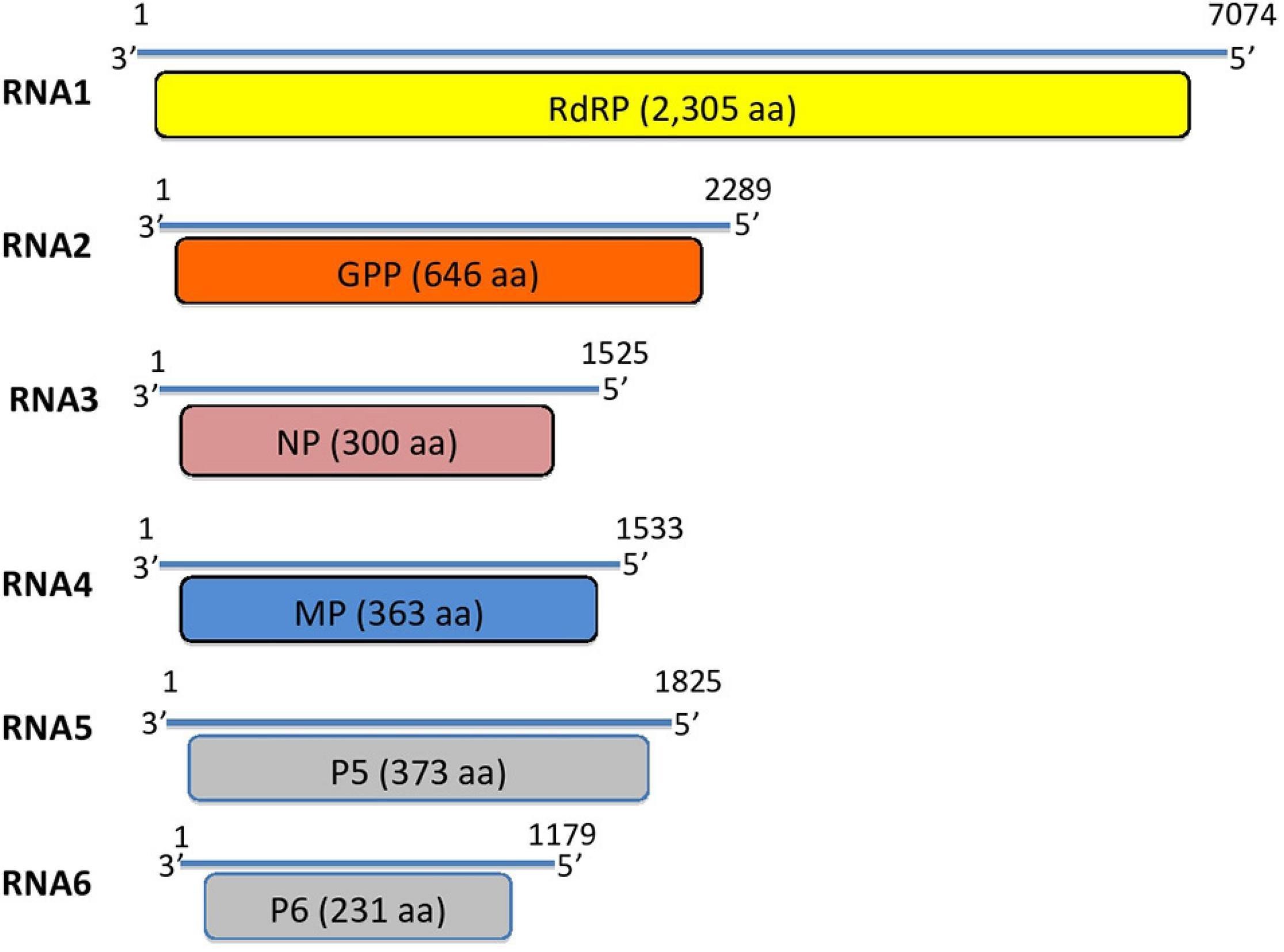
Figure 2. Genome segments of the novel maple mottle-associated virus (MaMaV). RdRP, RNA-dependent RNA polymerase; GPP, glycoprotein precursor; NP, nucleocapsid protein; MP, movement protein; P5, hypothetical protein; P6, hypothetical protein. Numbers represent the nucleotide positions of the RNA segments.
Further studies on symptomatic maple leaves collected in Berlin (Grunewald), Brandenburg (Forst Eberswalde and Melzower Forst), Bavaria (Laufen), and Bozen (Italy) revealed the presence of two additional emaraviruses genome segments related to MaMaV, which also occur as mixed infections in individual trees. The partial genomes of maple emaravirus 2 (MEV-2) (partial RNA1, RNA2, RNA3, RNA4) and maple emaravirus-3 (MEV-3) (RNA1, RNA2) have been determined by High Throughput Sequencing (HTS) and confirmed by RT-PCR in individual maple all showing typical symptoms of the mosaic disease. This suggests that at least three different emaraviruses are contributing to the mosaic disease of maple leading to the observed diversity and severity of leaf symptoms.
Transmission
Graft transmissibility of the mosaic disease has been proven by MaMaV-specific RT-PCR of virus-infected maple scions grafted onto rootstocks, developing mosaic and mottle symptoms in the first vegetation period (Hüttner, 2017).
Regarding transmission trough vectors, several members of the genus Emaravirus are known to be transmitted by eriophyid mites (Acari: Eriophyidae) (Mielke-Ehret and Mühlbach, 2012; Hassan et al., 2017; Elbeaino et al., 2018). In several of the diseased trees damages from gall mites (Aceria macrophylla, Eriophyes psilomerus) as well as from leafhoppers were found. The gall mites can be considered as putative vectors, but the hypothesis that they may be involved in the emaravirus transmission needs to be studied
-
-
-
-
Atanasoff, D., 1935. Old and new virus diseases of trees and shrubs. Phytopathol. Z. 8, 197–223.
-
Bandte, M., Hamacher, J., and Büttner, C., 2008. “Maple tree (Acer sp.) infected by viruses and phytoplasma,” in Jahrbuch der Baumpflege: Thalacker-Medien, eds H. D. Dujesiefken and P. Kockerbeck (Braunschweig: Deutsche Baumpflegetage), 208–212.
-
Büttner, C., von Bargen, S., and Bandte, M., 2015. “Chapter 13: phytopathogenic viruses,” in Principles of Plant Microbe Interactions: Role of Microbes in Sustainable Agriculture, ed. Lugtenberg (Heidelberg: Springer), 115–122. doi: 10.1007/978-3-319-08575-3_13.
-
Büttner, C., von Bargen, S., Bandte, M., and Muehlbach, H.P., 2013. “Forests diseases caused by viruses,” in Infectious Forest Diseases, eds P. Gonthier and G. Nicolotti (CABI), 50–75.
-
Cerescu D., 2022. Investigations of putative novel emaraviruses affecting maple (Acer pseudoplatanus). MSc-Arbeit, Humboldt-Universität zu Berlin, 119 Seiten.
-
Führling, M., and Büttner, C., 1998. Nachweis von Tobamo-Viren in Bergahorn (Acer pseudoplatanus L.) mit Scheckung und Blattdeformation. Forstw. Cbl. 117, 92–97. doi: 10.1007/bf02832962.
-
Hadidi, A., Barba, M., Candresse, T., and Jelkmann, W., 2011. Virus and Virus-Like Diseases of Pome and Stone Fruits. St. Paul, MN: APS Press.
-
Hüttner, F, 2017. Untersuchungen zur Pfropfübertragbarkeit von Emaraviren aus Populus tremula und Acer pseudoplatanus. BSc-Arbeit Humboldt-Universität zu Berlin, 65 Seiten.
-
Kaminska, M., and Suwa, H., 2006. First report of a decline of ash leaf maple (Acer negundo) in Poland, associated with Candidatus Phytoplasma asteris. Plant Pathol. 55:293. doi: 10.1111/j.1365-3059.2005.01304.x.
-
Massart, S., Candresse, T., Gil, J., Lacomme, C., Predajna, L., Ravnikar, M., et al., 2017. A framework for the evaluation of biosecurity, commercial, regulatory and scientific impacts of plant viruses and viroids identified by NGS technologies. Front. Microbiol. 8:45. doi: 10.3389/fmicb.2017.00045.
-
Mielke-Ehret, N., and Mühlbach, H.-P., 2012. Emaravirus: a novel genus of multipartite, negative strand RNA plant viruses. Viruses 4, 1515–1536. doi: 10.3390/v4091515.
-
Rehanek M., Karlin D.G., Bandte M., Al Kubrusli R., Nourinejhad Zarghani S., Candresse T., Büttner C., von Bargen S., 2022. The Complex World of Emaraviruses—Challenges, Insights and Prospects. Forests 13, 1868. [https://doi.org/10.3390/ f13111868].
-
Rumbou, A.; Candresse, T.; von Bargen, S.; Büttner, C., 2021. Next-generation sequencing reveals a novel emaravirus in diseased maple trees from a German urban forest. Front. Microbiol., 11, 621179. Doi: 10.3390/microorganisms9081730.
-
Rumbou, A.; Vainio, E.J.; Büttner, C., 2021. Towards the Forest Virome: High-Throughput Sequencing Drastically Expands Our Understanding on Virosphere in Temperate Forest Ecosystems. Microorganisms, 9, 1730. https://doi.org/10.3390/microorganisms9081730.
-
Tatineni, S., McMechan, A. J., Wosula, E. N., Wegulo, S. N., Graybosch, R. A., French, R., et al., 2014. An eriophyid mite-transmitted plant virus contains eight genomic RNA segments with unusual heterogeneity in the nucleocapsid protein. J. Virol. 88, 11834–11845. doi: 10.1128/jvi.01901-14.
-
Wulf, A., and Kehr, R., 2009. “Diseases, disorders and pests of selected valuable broadleaved tree species,” in Valuable Broadleaved Forests in Europe, eds H. Spiecker, S. Hein, K. Makkonen-Spiecker, and M. Thies (Leiden: Brill), 61–84.
-
Vainio, E.J., Rumbou, A., Diez, J.J., Büttner, C., 2024. Forest Tree Virome as a Source of Tree Diseases and Biological Control Agents. Curr. For. Rep. 10, 153–174. https://doi.org/10.1007/s40725-024-00214-8.
-
-
-